 VOLVE 5.0
VOLVE 5.0
Organisms and Cells
This page will discusss the mechanics of how cells and organisms interact with the universe.
Theory of Operation
CELLs are CPUs that have a small repertoire of instructions for manipulating the world. All of the physics of the simulation are embodied by these instructions. The general physics to be maintained are: (1) Locality (most instructions operate on nearest neighbors). (2) Conservation of energy (energy is only ever transferred). (3) Exclusion principle (nothing can occupy the same grid location as something else). (4) Connectivity of the cells in an organism.
These instructions can all be independenly enabled/disabled in your simulation. Each instruction can be tweaked and modified using mode flags.
These instructions need a programmatic way to specify what they want to happen. The most important detail is specifying (x,y) coordinates for the instruction. So coordinates will be discussed next.
Table of Contents:
- Direction Vectors
- Normalized Vectors
- Organism Connectivity
- Energy
- Instruction: OMOVE
- Instruction: ROTATE
- Instruction: CMOVE
- Instruction: CSHIFT
- Instruction: GROW
- Instruction: MAKE-SPORE
- Instruction: EAT
- Instruction: LOOK
- Instruction: NEAREST
- Instruction: FARTHEST
- Instructions: SMELL, EXUDE
- Communicating Between Cells
- Instructions: READ, WRITE
- Organism Instruction Reference
Coordinate System
The grid uses the following (x,y) coordinate system:
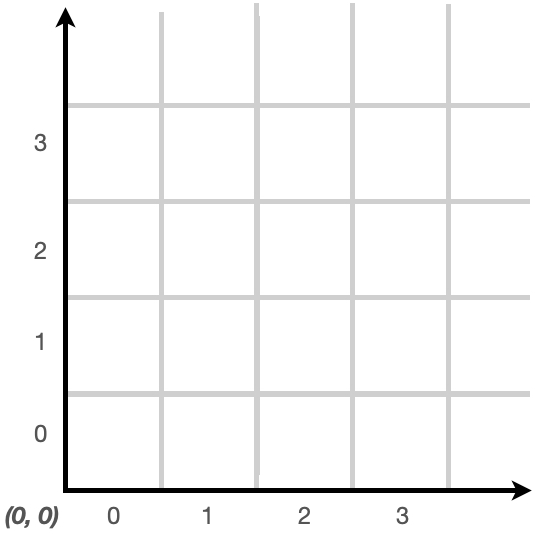
Here's a simulation screen showing the origin in the lower left.
Directions
Many of the instructions that cells execute require a vector. This vector is used to indicate a direction, or an adjacent square that the organism or cell will interact with. This diagram shows (x, y) coordinates for the eight directions surrounding a cell:
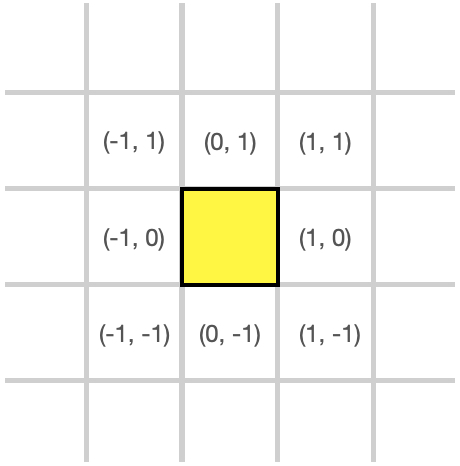
Most operations that interact with the universe using (x, y) coordinates will specify coordinates like these. This is called a normalized direction vector.
Not shown is the direction (0, 0) which correponds to the cell in the middle. This is normally the cell executing the instruction.
Some instructions use non-normalized direction vectors, such as MOOD and SEND. In this case any values may be given for (x, y) such as (-4, 2).
Normalized Vectors
Because most operations are restricted to the adjacent squares, (x,y) vectors will often be normalized before using them. Here is how Evolve instructions normalize coordinates to values of -1, 0, +1.- values less than 0 (< 0) => map to -1.
- values equal to 0 (= 0) => map to 0.
- values more than 0 (> 0) => map to 1.
For example,
-30 4 OMOVE ; move organism using vector (-1, 1) 0 -4 CMOVE ; move cell using vector (0, -1) 9 29 5 MAKE-SPORE ; make spore using vector (1, 1) (with energy 5)
Organism Connectivity
Organisms consists of one or more cells. To ensure the shape of an organism remains coherent Evolve will enforce the connectivity constraint that a cell much be reachable in one of the 8 directions surrounding the cell.
Here are examples of organisms:
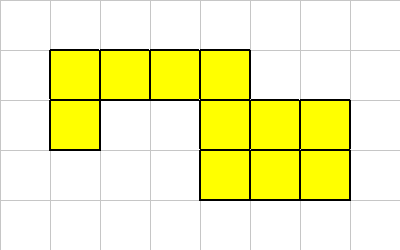
One of its corner cells has been killed,
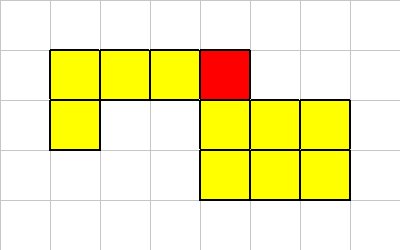
Our connectivity constraints are such that a multi-cellular organism must always have at least one neighbor in one of the 8 directions.
In this example, after the dead cell is removed, we still have a properly connected organism.
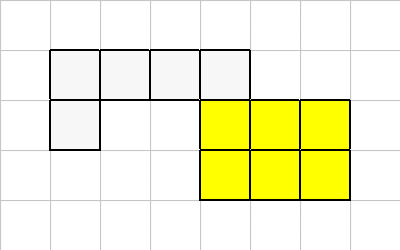
Sometimes the connectivity between the cells is broken. If a cell is kills, the largest region is kept, and the smaller regions will be converted to organic material.
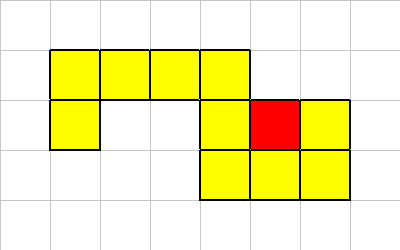
In this case the death of this cell does not break the organism into multiple sections. So in this case we just kill of the cell in question.
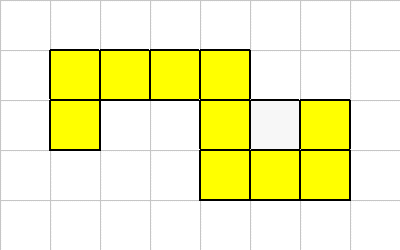
These are properly connected:
This is because all the cells are touching in at least one of the 8 directions.
Energy
Energy is an attribute of each organism. The total amount of energy in a simulation is equal to the amount of energy that the first organism(s) had when the simulation was created. The total energy never changes during a simulation (conservation of energy).
Every cell must have at least one unit of energy just to be considered "alive". If some operation causes this value to go to zero, the cell is dead.
Why Energy?
The purpose of energy is to introduce a limit on organism/cell growth. Without it, organisms would replicate without limit. Other constrains may be introduced that changes how energy is used to limit growth.
Energy and Organic Material
When a cell is killed by being eaten, the connectivity algorithm must analyse the organism and kill of all but the largest region. Each cell in a region that is killed off will be converted to organic material. The amount of energy that this organic material will contain is equal to 1/n th the amount of energy in the organism (where n is the number of cells in the organism) plus any stack elements beyond 10.
Energy and Spores
A spore gets created with an energy amount (specified by the MAKE-SPORE instruction). If a spore gets eaten then this energy amount is transferred to the organism that ate it. If a spore is fertilized, then the new organism gets created with the sum of energy from the first and second MAKE-SPORE instructions.
Energy and Other Operations
Energy is not needed to move, look, eat, etc.... Energy is only needed to Grow, Make a spore, Reproduce.
Use energy to control the overall population of your simulation:
- 100,000 units of energy equals about 1,000 organisms.
- 500,000 units of energy equals about 2,000 - 3,000 organisms.
- 1,000,000 units of energy equals about 5,000 - 8,000 organisms.
Other things will determine the population you will see. Energy Requirements are configurable on the Modes tab.
OMOVE
The OMOVE instruction causes the entire organism to move in one of the 8 directions.
Assume that cell 2 has executed the instruction 1 0 OMOVE. The vector (1,0) means move right 1 square.
Or assume that cell 1 has executed the instruction -1 -1 OMOVE. The vector (-1, -1) means move left and down diagonally.
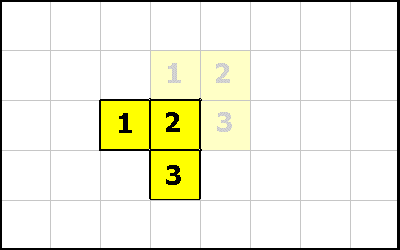
The OMOVE instruction first checks that all squares that organism will move to, are vacant. Note that any cell could have executed these instructions and the organism would have moved the same.
ROTATE
The ROTATE instruction causes the organism to be rotated. Because of the grid layout and the constraints on the shape of organisms, this instruction only rotates in 90 (or 45) degree units. Rotate takes a single integer off of the data stack. This value represents 90 (or 45) degree rotations.
Consider this organism, the cell with the dot will execute a ROTATE instruction. (NOTE: In these examples the ROTATE mode is confugured for 90 degree rotations, and the center of rotation is the cell executing the instruction. These properties can be changed by changing the mode bits.)
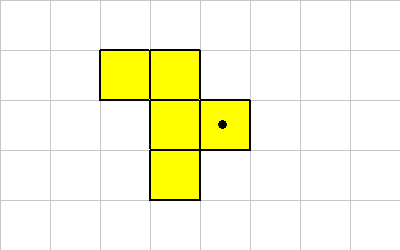
Here's what the organism looks like after a -1 ROTATE:
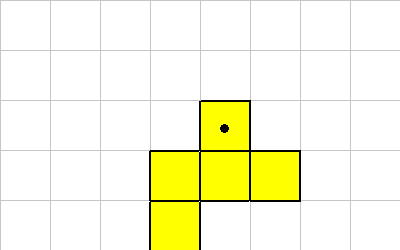
Here's what the organism looks like after a 1 ROTATE (from starting figure):
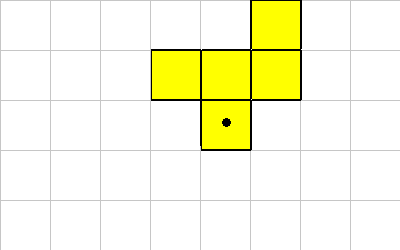
This instruction requires that all grid locations that the rotated organism is to be moved are vacant.
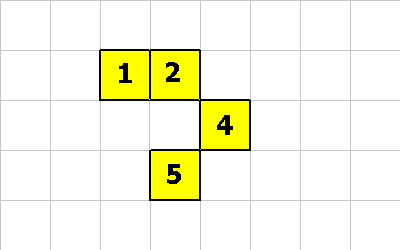
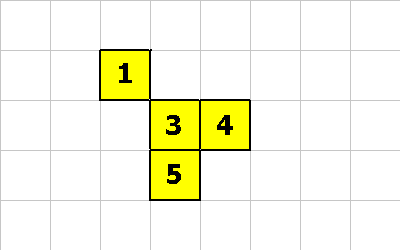
CMOVE
CMOVE is a cool instruction. It allows a cell to move relative to the organism. The following pictures will demonstrate an organism morphing from one shape to another. The following instructions will be executed:
0 -1 CMOVE ; cell 1 executes this instruction 0 -1 CMOVE ; then cell 4 executes this instruction 1 1 CMOVE ; then cell 5 executes this instruction
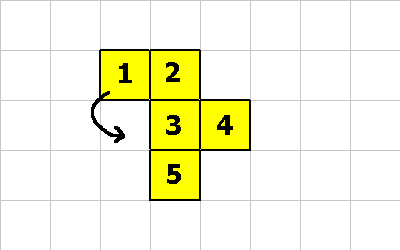
0 -1 CMOVE ; <--- cell 1 executes this
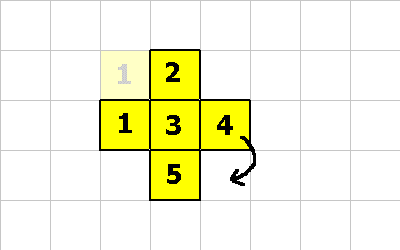
0 -1 CMOVE ; <--- cell 4 executes this
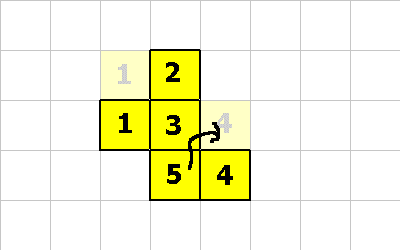
1 1 CMOVE ; <--- cell 5 executes this
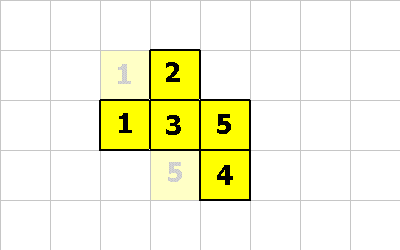
This last pictures shows what the creature looks like after these 3 CMOVE instructions.
For the the CMOVE operation to succeed the shape being morphed into must always be properly connected.
CSHIFT
The CSHIFT instruction allows the shape of an organism to change by shifting cells along a line. As long as the final shape is properly connected the CSHIFT instruction is allowed. Here is the initial state of some organism. We will see it transform after executing three CSHIFT instructions.
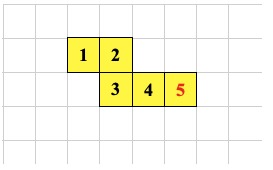
-1 0 CSHIFT ; <--- cell 5 executes this
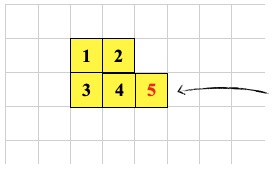
-1 0 CSHIFT ; <--- cell 5 executes this a second time
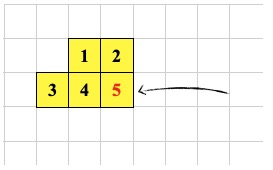
0 -1 CSHIFT ; <--- cell 2 executes this
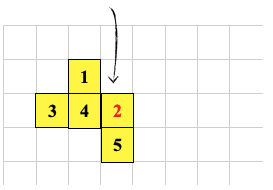
GROW
The GROW instruction is how an organism (which always starts out life as a single cell) becoms multi-cellular.
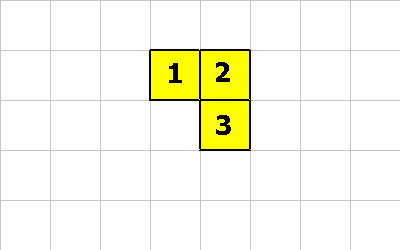
Let's assume this is our organism:
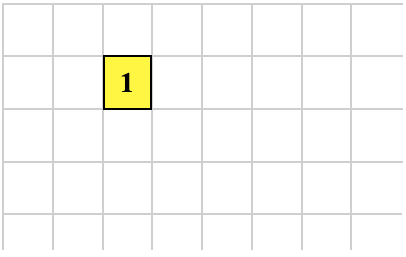
To get to the state shown below cell 1 executes this:
1 0 GROW ; executed by cell 1
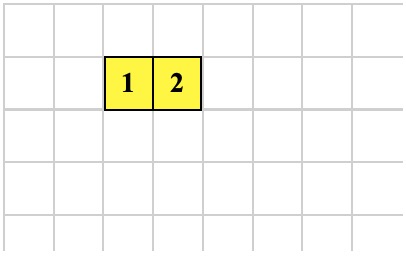
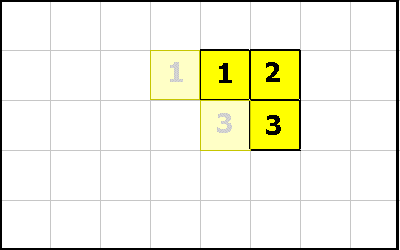
Another GROW operation produces the organism below. What are the possible operations that could create the organism? Here is the only possibility:
1 -1 GROW ; executed by cell 2
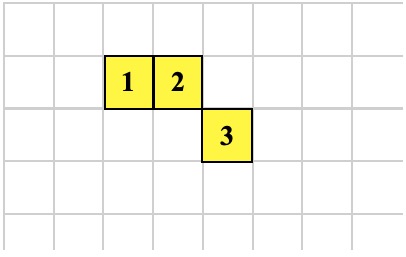
As shown above, the GROW operation creates a new cell. The new cell recieves the execution context of the cell that executed the GROW instruction. The only difference is the new cell has a -1 on top of its data stack and the other cell has a 1.
Grow is very much like the fork() system call on Unix. Fork creates a new process. But fork does it by cloning the current process. The only way for a programmer to know which execution context they are in is by examining the return value of fork(). The same applies to the GROW instruction.
Grow also allows a creature to increase its processing power. An organism consisting of two cells has twice the processing power of a single celled creature. Such an organism can do twice the computations in the same amount of steps as a single celled creature. This is profound capability and any creature that makes the leap into multi-cellularism has a lot going for it.
Example KFORTH code using GROW,
main:
{
1 0 GROW -1 = LeafCell if
brain call
}
LeafCell:
{
1 0 EAT pop
-1 -1 EAT pop
1 ?loop
}
brain:
{
; ... creature brain goes here ...
}
|
In this example a new cell is grown at coordinates (1, 0), and the new cell begins its life executing inside of the code block 'LeafCell'. The LeafCell just eat's forever. The parent cell continues to call the 'brain' routine to do the normal creature processing.
You can write your routine to encapsulate this common logic as follows:
main:
{
1 0 LeafCell myGROW call
brain call
}
LeafCell:
{
; .... leaf cell stuff here ...
}
brain:
{
; ... brain cell stuff here ...
}
;
; (x y cb -- )
;
; Grow and call 'cb' for the new cell
;
myGROW:
{
-rot GROW -1 = { pop } ifelse
}
|
MAKE-SPORE
The MAKE-SPORE instruction is how reproduction occurs. In order to reproduce a spore must be created. But this isn't enough. Another spore must be created on top of the first one.
This mechanism allows both sexual and asexual reproduction. (see spores for more information). The following example shows an organism reproducing asexually:
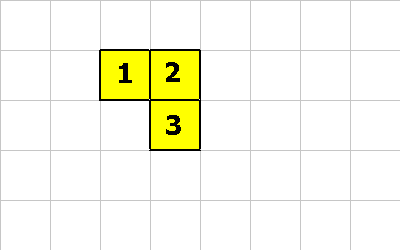
So our happy organism wants to have a baby. Either cell 2 executes this instruction:
1 0 20 MAKE-SPORE ; create spore with 20 units of energy
Or cell 3 executes this one:
1 1 20 MAKE-SPORE ; create spore with 20 units of energy
In either case this is what the universe now looks like:
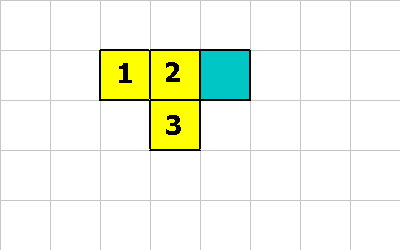
Now lets assume one of the following instructions gets executed:
1 0 12 MAKE-SPORE ; cell 2 does this or.... 1 -1 12 MAKE-SPORE ; cell 3 does this
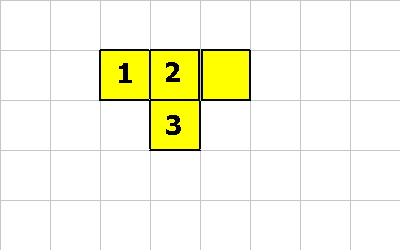
A new organism is born! Whooo hoo. The new organism will start out life with 32 units of energy (20 + 12). The new organism will inherit the genetic program of the parent organism. (with mutations).
Sexual reproduction occurs when two different organisms contribute a spore. In this case the genetic program from both parents is interlaced (this process is described elsewhere).
All organisms belong to a certain strain (by default strain 0). Strains allow you to tag a lineage of organisms from the first one to an evolved population. Therefore, SPORE's cannot be fertilized if the two organisms belong to different strains.
The first MAKE-SPORE operation must have an energy amount that is greather than 0. But the second MAKE-SPORE instruction can fertilize an existing spore using 0 energy.
EAT
Eating is how an organism survives. The EAT instruction must be called to eat stuff that is touching the cell. The cell executing the EAT instruction will attempt to eat anything at the (x, y) offset.
EAT'ing is basically the transfer of energy between cells. Related to this is the SEND-ENERGY instruction. It is like EAT except it allows a precise amount of energy to be transferred. SEND-ENERGY can be used when inventing ones own simulation rules.
In this example cell 2, executes the 1 -1 EAT instruction.
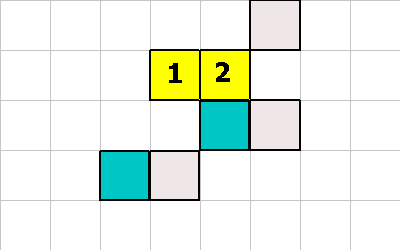
After the instruction the white square (organic material) has been removed.
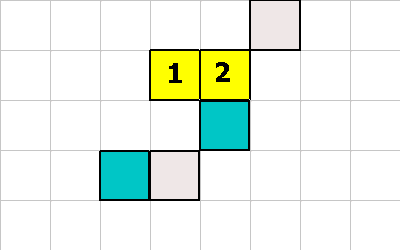
Not shown is what happens when a cell tries to eat another living cell. In this case the cell being eaten is flagged as dead (it will show up RED). The organism which has dead cells will have to resolve that issue later in the simulation.
The precise rules for EAT and SEND-ENERGY are given by mode flags for EAT and SEND-ENERGY.
To eat the spore located at (0, -1) this instruction could be used: 0 -1 EAT.
LOOK
The LOOK instruction takes a normalized (x, y) vector and looks along that line until it finds something. The thing it finds (the 'what' value) and the distance in which it was found it returned on the data stack.
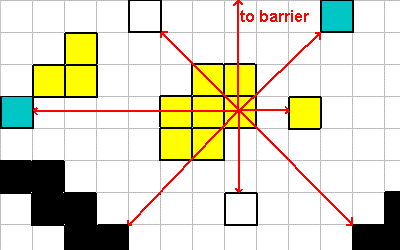
This figure shows all 8 direction vectors from a single cell. The LOOK instruction would return the following for this cell:
0 1 LOOK ; what = 8, where = 100 1 1 LOOK ; what = 2, where = 3 1 0 LOOK ; what = 1, where = 2 1 -1 LOOK ; what = 8, where = 4 0 -1 LOOK ; what = 4, where = 3 -1 -1 LOOK ; what = 8, where = 4 -1 0 LOOK ; what = 2, where = 7 -1 1 LOOK ; what = 4, where = 3 0 0 LOOK ; what = 0, where = 0
Vision using LOOK (and the other vision instructions) will look through the organism itself. This makes the vision instructions less evolutionary brittle, as an "eye" cell can evolve inside the middle of a multi-cellular organism.
The 'what' values are powers of 2, which allows them to be OR'd together for form a search mask (see NEAREST and FARTHEST).
A LOOK operation using the vector (0, 0) returns a what=0, where=0.
"What" values are assigned according to this key:
- 1 = Cell
- 2 = Spore
- 4 = Organic Material
- 8 = Barrier
When LOOK is configured to not be able to see through itself, then the additional bit 16 will be set. Also LOOK can be configuired to reveal strain bits. Here is the addtional bits that can be returned by LOOK (and the other vision instructions).
- 1 = Cell
- 2 = Spore
- 4 = Organic Material
- 8 = Barrier
- 16 = Self
- 32 = Strain 0
- 64 = Strain 1
- 128 = Strain 2
- 256 = Strain 3
- 512 = Strain 4
- 1024 = Strain 5
- 2048 = Strain 6
- 4096 = Strain 7
NEAREST
This instruction looks in all 8 directions surrounding a cell and returns the (x, y) vector that has the smallest distance.
This instruction takes a bit-mask which is an OR'ing of 'what' values. Only these types of things will be used when determining the nearest direction.
For example, a mask value of 5 (binary 0101), matches Cells and Organics (but not spores or barriers). So this code,

5 NEAREST ; returns (2, 0) |
Would return vector (1, 0) because that direction has a "cell" at distance 2 (which is the smallest distance that matches the bit-mask).
For all vision instructions which take a 'what' mask, the matching criteria works as follows: We AND the two what values together. If non-zero then the object matched. If zero we stop searching along the line and report 0 0. This allows filtering for certain strains as well for certain types of blocks.
NOTE: If more than one direction "tie" for seeing the nearest (or farthest) thing, then instructions NEAREST, FARTHEST, BIGGEST, SMALLEST, HOTTEST, COLDEST will resolve the tie by choosing one of the matches randomly.
NOTE 2: The returned vector from these instructions are not normalized. This means they return the distance as well as the direction. For example (-6, -6) is returned instead of (-1, -1) to indicate both the distance and direction. The DIST instruction can be applied to this vector to get the distance, 6.
FARTHEST
This instruction looks in all 8 directions surrounding a cell and returns the (x, y) vector that has the largest distance.
This instruction takes a bit-mask which is an OR'ing of 'what' values. Only these types of things will be used when determining the farthest direction.
For example, a mask value of 2 (binary 0010), matches only spores (but nothing else). So this code,

2 FARTHEST ; returns (-7, 0) |
Would return vector (-7, 0) because that direction has a "spore" at distance 7 (which is the largest distance to any spore).
SMELL
The grid contains an odor value at each (x,y) coordinate. These instructions allow for this 16-bit value to be read/written. The smell instruction lets the cell read a value from the grid.
1 -1 SMELL ; retrieves the value 2 from the grid |
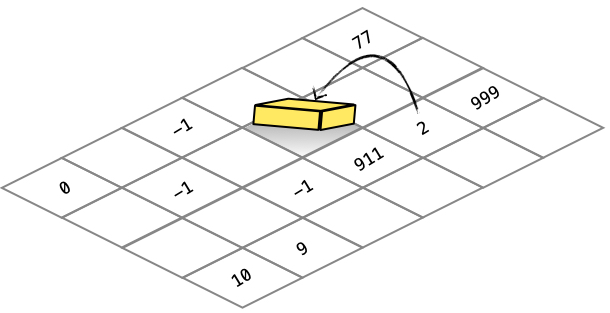
EXUDE
The exude instruction allows the cell to write a value to the grid.
1 -1 SMELL ; retrieve the value 2 from the grid 125 - ; 2 - 125 = -123 -1 0 EXUDE ; writes the value -123 to the grid |
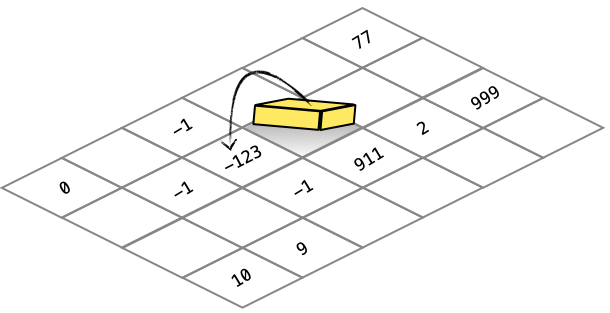
The SMELL and EXUDE instructions use normalized direction vectors to read and write the odor map values from the surrounding 9 squares.
Communicating Between Cells
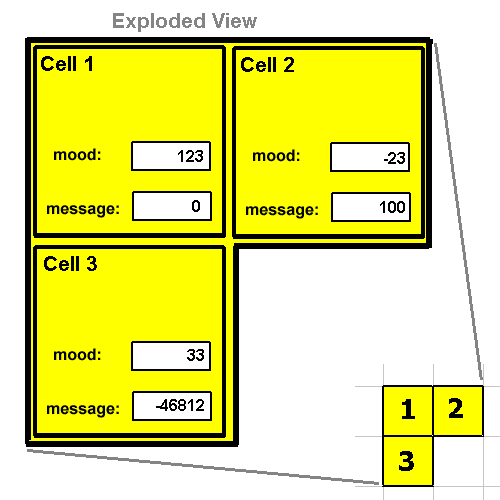
Refer to this figure for the descriptions of the following instructions. The mood register is something the cell sets, and others can look at. The message register is something that others set, and this cell looks at.
SEND
Using the instructions pack2, unpack2 two values can be combined into a single 16-bit integer and stored in the MOOD and MESSAGE registers. For example, to send 2 integers to another cell, this code could be used:
23 -99 pack2 ; combined 2 (8-bit integers) into single 16-bit int. -1 1 SEND ; send to cell |
RECV
The recieving cell can extract the 2 values like so:
RECV unpack2 ; the values 23, -99 are now on the stack |
BROADCAST
This instruction broadcasts the same value to all cells in the organism.
3200 BROADCAST ; put 3200 in everybody's MESSAGE register |
SAY
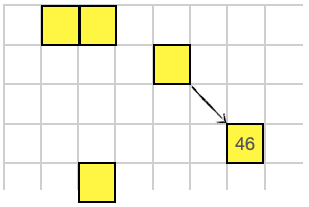
The SAY instruction will send a message (a single 16-bit value) to another organism. The recieving organism uses the RECV instruction to get the message. The message is sent along the noramlize direction vector to the firs thing it encounters.
46 1 -1 SAY
LISTEN
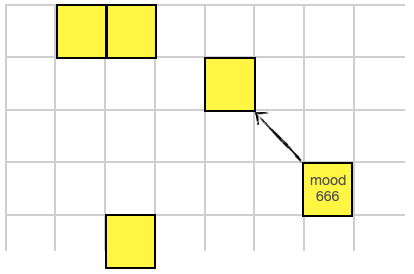
The LISTEN instruction will retrieve the MOOD register (a single 16-bit value) from another cell. The mood value is "heard" along the noramlize direction vector given by the (x,y) values on the data stack. The value of that other cell's MOOD register is put on this cells data stack.
1 -1 LISTEN ; retrieves the value 666
SHOUT
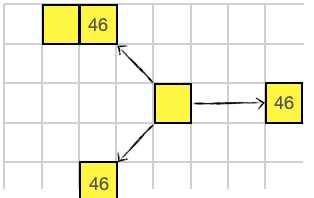
Why speak when you can shout? The SHOUT instruction broadcasts a message (a single 16-bit value) along all 8 directions. Any cell in line will recieve this message. The recieving organism uses the RECV instruction to get the message.
46 SHOUT
Interrupts
These communications instructions can be configured to interrupt the cell upon recieving a message. An interrupt uses the TRAP handlers to transfer control to a protected code block. See the mode flags for these instructions: SEND, BROADCAST, EAT, SEND-ENERGY, SHOUT, SAY, WRITE, KEY-PRESS.
WRITE
The WRITE instruction allows a cell to write data to another organism or spore (a spore is shown here).
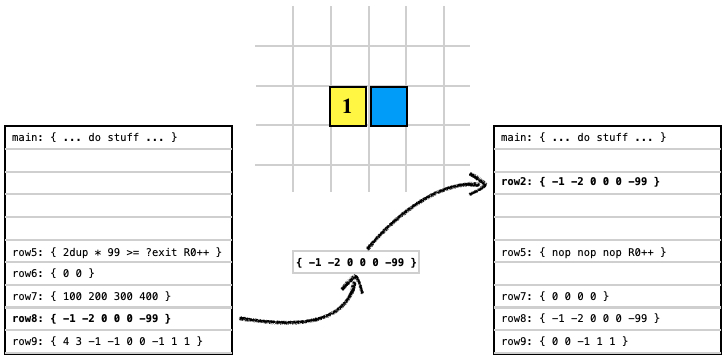
0 1 2 8 WRITE pop ; write code block 8 to spore at (0,1) code block 2 |
READ
The READ instruction allows a cell to read data from another organism or spore (a spore is shown here).
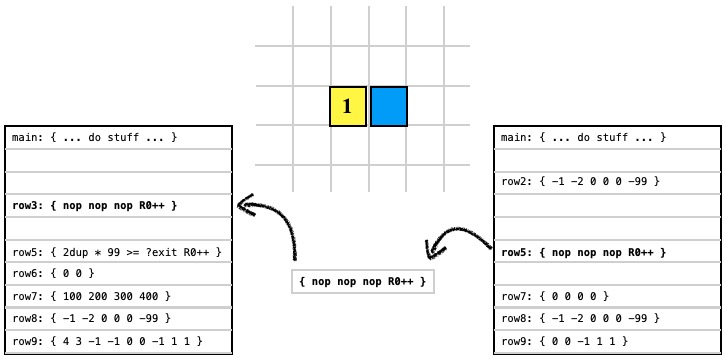
0 1 5 3 READ ; read code block 5 from spore at (0,1) put data in code block 3 |
These instructions allows organism to communicate large amounts of data between each other. Also, spores can be used as storage devices to store and retrieve "files" of data. Note that these instructions can exchange code as well as data.
The READ/WRITE instructions give the simulator more core wars style parasitism and other tricks. Genetic information can be spread in this way without using the normal replication instructions.
The purpose of the READ/WRITE instructions is to fill out the computational model of Evolve 5.0 universe. It allows data to be exchanged akin to files or records in an operating system. I like to think of spores as "files" that you can place on the grid, much like an operating system lets you place files in the file system.
ORGANISM INSTRUCTION REFERENCE
The core KFORTH instruction (like dup, pop, +, *, /, etc..) are covered in the section entitled the KFORTH Language. What follows is a detailed reference guide for the instructions that control organisms and cells:
Interacting with the universe:
Vision:
Communicating between cells:
Query information about ourselves:
Universe Instructions:
Misc. Instructions:
CMOVE
Usage: ( x y -- r )move a cell relative to the organism
Move the cell that executes this instruction relative to the organism.
- If there are not 2 elements on the data stack, then skip this instruction.
- Remove the (x, y) coordinates from the stack.
- Normalize the coordinates.
- Examine the destination square (as indicated by the offsets) and see if it is vacant. If not, return 0.
- Check that the organism is propery connected after the move. If not return 0.
- Else, Move the cell to the new location and return 1.
- A CMOVE of (0,0) always fails and returns 0.
RETURNS:
- 0 - cell could not be moved.
- 1 - success.
ENERGY: Does not require any energy.
CMOVE MODE BITS:
| BIT | MASK | Meaning when bit is 0 | Meaning when bit is 1 |
| 0 | 1 | NOT USED | NOT USED |
move entire organism Move entire organism by one square in the up, down, left, right, or diagonal direction. RETURNS: ENERGY: Does not require any energy. OMOVE MODE BITS: rotate organism Rotate the organism by 45 degree units around the cell that executes this instruction. The rotate direction is given by
'n'. A positive 'n' rotates the organism clock-wise 45 degrees. A negative 'n' rotates the organism 45 degrees counter-clockwise.
If 'n' is zero don't rotate the organism and return 0.
RETURNS: ENERGY: Does not require any energy. ROTATE MODE BITS: eat energy from surrounding cells Eat stuff at (x, y) from this cell. Return 'n' the amount of energy eaten. RETURNS: ENERGY: Does not require any energy. EAT MODE BITS: give/take energy to neighboring cell send the energy to the cell at normalized coordinate (x,y). negative 'e' values take from that cell. If there isn't
enough energy to perform the transfer (too little energy compared to 'e')) then don't do anything. RETURNS: Returns the number of units of energy given/taken. 0 is returned if the operation failed. ENERGY: SEND-ENERGY MODE BITS: create spore and reproduce Create new spore (or fertilize an existing spore).
When a spore is fertilized, the original genetic program used to create the spore, and the genetic program from the
organism that fertilized the spore will be first merged. Then the resulting genetic program is MUTATED. Then a new organism
is born. It will begin execution at the first instruction of code block 0 (main). It will be given 'e' units of energy, which is the sum of the energy specified with the first and second MAKE-SPORE instructions.
MAKE-SPORE Energy: This setting is available on the Strain Modes tab. The value represents the minimum amount of energy required to
create a spore. If 'e' is not big enough, then this instruction will return 0. A minimum of '1' units
of energy is always required to create the initial spore.
RETURNS: ENERGY: Requires 'e' units of energy. MAKE-SPORE MODE BITS: add a new cell to the organism
Add a new cell to this organism. The new cell inherits the data stack, call stack and registers of the
parent cell. The parent and child can check the return value to determine which one they are.
GROW Energy: This field is available on the Strain Modes tab. It sets the minimum amount of energy required to grow
a cell. Each organism has a total amount of energy. If total energy divided by the number of cells isn't
equal or greater to this value, then the GROW operation will fail.
GROW Max Size: This field is available on the Strain Modes tab. It sets the maximum number of cells that an organism
can be. If this value is 0, then there is no maximum.
RETURNS: ENERGY: Requires GROW Energy units of energy. GROW MODE BITS: create organic block Create organic block at (x,y) offset with energy e. RETURNS: 0 Returns the amount of energy that was d ENERGY: The energy 'e' is deducted from the organisms (or cells) energy. MAKE-ORGANIC MODE BITS: grow another cell Grow a new cell at (x,y) normalized offset. The new cell inherits the context of the cell which executed the
GROW.CB instruction. However, it will begin execution at code block 'cb'. Respects the Grow Max Size setting. If the number of cells
is already at the max size, then GROW.CB will not succeed. Unprotected code cannot specify a protected code block number
for cb. RETURNS: ENERGY: Respects the Grow Energy setting. If adding this cell would cause the total amount of energy per cell to drop below
the Grow Energy amount, then GROW.CB fails. shift a line of cells relative to the organism This instruction is similar to CMOVE except a group of cells move (or shift position in space) along a vector.
If the shape of the organism after the CSHIFT operation would violate the connectivity of the organism then this instruction
fails. RETURNS: Returns the number of cells that shifted. ENERGY: Does not require energy. CSHIFT MODE BITS: spawn a new organism of a particular strain Spawn a new organism at the (x,y) normalized offset. Give it 'e' units of energy. Start running at code block
'cb. The identity of the new organism will be strain 'strain'. The cellular state of the spawned organism will inherit
the cell executing this instruction. Unprotected code cannot specify a protected code block number for cb. RETURNS: returns 1 if the spawn was successful, else 0. ENERGY:'e' units of energy are required. SPAWN MODE BITS: create (or clear) a barrier block Creates a barrier block at the x y normalized offet. If the location already contains a barrier block, then this
instruction will clear the barrier. RETURNS: ENERGY: MAKE-BARRIER MODE BITS: excrete a value onto the grid excrete a value onto the grid RETURNS: ENERGY: EXUDE MODE BITS: read a value from the grid read a value from the grid that was previously EXUDE'd. RETURNS: ENERGY: look along a line Look along a normalized (x, y) direction vector. Return the thing it found, and the distance.
NOTE: The LOOK Mode settings apply to all the vision instructions such as NEAREST / FARTHEST, HOTTEST / COLDEST, and SMALLEST
/ BIGGEST.
RETURNS:
What values:
ENERGY: Does not use energy. LOOK MODE BITS: find nearest object Looks in all 8 directions and return the (x, y) direction vector that corresponds to the thing with the smallest
distance (and matches a 'mask' value). The returned (x,y) vector is not normalized, so it might return (-6,6) instead of
(-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
RETURNS: ENERGY: Does not use energy. find farthest object Looks in all 8 directions and return the (x, y) direction vector that corresponds to the thing with the largest
distance (and matches a 'mask' value). The returned (x,y) vector is not normalized, so it might return (-6,6) instead of
(-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
RETURNS: ENERGY: Does not use energy. see the size of something Report the size (number of cells) of the object seen along the normalized vector (x,y) from this cell. RETURNS: The number of cells (the 'size') in the organism seen along (x,y). Also returns the distance. ENERGY: find the biggest thing in view Look in all 8 directions. Return a non-normalized (x,y) vector to the biggest thing. This is the organism consisting of
the most number of cells. The returned (x,y) vector is not normalized, so it might return (-6,6) instead of (-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
This instruction also uses the LOOK Mode bits.
RETURNS: ENERGY: find the smallest thing in view Look in all 8 directions. Return a non-normalized (x,y) vector to the smallest thing. This is the organism with the
least number of cells. The returned (x,y) vector is not normalized, so it might return (-6,6) instead of (-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
This instruction also uses the LOOK Mode bits.
RETURNS: ENERGY: get the energy of cell seen along (x,y) Get the energy of cell seen along (x,y). This is per cell. For organisms we divide the total energy the
organism has by the number of cells the organism has. RETURNS: ENERGY: find the most energetic cell in view Look in all 8 directions. Return a non-normalized (x,y) vector to the hottest thing. This is the organism with the
most energy per cell. The returned (x,y) vector is not normalized, so it might return (-6,6) instead of (-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
This instruction also uses the LOOK Mode bits.
RETURNS: ENERGY: find the least energetic cell in view Look in all 8 directions. Return a non-normalized (x,y) vector to the coldest thing. This is the organism with the
least energy per cell. The returned (x,y) vector is not normalized, so it might return (-6,6) instead of (-1,1).
mask values are the same as what values. If any bit matches the what from LOOK'ing along that vector then
we match.
This instruction also uses the LOOK Mode bits.
RETURNS: ENERGY: Query the MOOD of one of my cells
Every cell has its own "MOOD" register. This instruction queries another cell using an (x, y) vector. This vector is not normalized which allows the cell to query any other cell within its own organism (not just adjacent cells). For example, if
cell 2 executed the following instructions:
If a bogus (x, y) is specified then 0 is pushed on the data stack. We can query our own
mood by specifying (0, 0).
RETURNS: ENERGY: Does not use energy. Set MOOD register to a value
The cell executing this instruction will set its own mood register. It is not possible to set others. The purpose
of the mood register is for a cell it advertise its "mood" to other cells in the organism. For example,
if cell 3 executed these instructions:
Then cell 3's mood would be increased by 1. So it would be set to 34.
RETURNS: ENERGY: Does not use energy. broadcast a message to our cells This instruction takes the value off of the top of the data stack and sets the message register for every
cell in our organism. If interrupts are enabled every cell, except the cell executing this instruction, will be interrupted. RETURNS: ENERGY: Does not use energy. BROADCAST MODE BITS: Set the MESSAGE register of one of my cells
This instruction takes 3 arguments. An (x, y) vector and a value 'm'. The vector is NOT normalized so that the cell
can send a message 'm' to any other cell in the same organism (not just adjacent cells). For example, if
cell 1 exectuted this these two instructions:
A SEND operation using bogus coordinates will be ignored.
RETURNS: ENERGY: Does not use energy. SEND MODE BITS: recieve message The RECV instruction just pushes a copy of our message register on top of the data stack. RETURNS: ENERGY: Does not use energy. broadcast a message to another organism Broadcast a message to another organism. Send the message 'm' outward in all 8 directions. If a cell is reached,
then set it's MESSAGE register to be 'm'. RETURNS: The number of cells that recieved the message. ENERGY: SHOUT MODE BITS: send a message along a line to another organism The (x,y) normalized direction vector will be used to send the message 'm'. If a cell is reached, then set
it's MESSAGE register to be 'm'. RETURNS: Returns non-zero on success. The returned value is the distance to the cell that successfully recieved the message, or 0
if no message delivered. ENERGY: SAY MODE BITS: read MOOD register from a cell from another organism Listen along the normalized vector (x,y). If another cell was found return its MOOD register and distance. RETURNS: Returns the mood and distance value, or 0 0 if nothing heard. ENERGY: LISTEN MODE BITS: read an entire code block from another cell or spore Read code block 'cb' from the cell or spore located at the normalized (x,y) coordinates. Place the result into 'cbme'.
It is not possible to grow the number of code blocks of a program with this instruction. The destination code
block 'cbme' must already exist.
Unprotected code cannot READ if 'cb' or 'cbme' is protected. Unprotected code cannot read a protected instruction.
RETURNS: On success, the length of the code block read is returned. On failure, a negative error code is returned. ENERGY: READ MODE BITS: write an entire code block to another cell or spore Write code block 'cbme' to another cell or spore. The cell is located at the normalized (x, y) offset from
this cell. The destination code block number is given by 'cb'. It is not possible to grow the number of
code blocks of a program. The destination code block 'cb' must already exist.
Unprotected code cannot WRITE if 'cb' or 'cbme' is protected. Unprotected code cannot write a protected instruction.
RETURNS: On success, the length of the code block written is returned. On failure, a negative error code is returned. ENERGY: WRITE MODE BITS: get energy of organism Get our organisms overall energy. RETURNS: ENERGY: return age of organism Fetch the organisms "age" field (which is the number of elapsed simulation step) and push this value on our data
stack. RETURNS:
The age of the organism
ENERGY: Does not use energy. number of cells Push the number of cells that comprise this organism on our data stack. RETURNS:
number of cells in this organism
ENERGY: Does not use energy. check for our cells Determine if a neighboring cell location contains our cell or not. RETURNS: ENERGY: Does not use energy. Returns a bitmask with a '1' bit set for a neighor in that direction. RETURNS: A bit mask representing the surrounding neighbors. ENERGY: get current position Returns the absolute grid coordinates for the cell executing this instruction. RETURNS: ENERGY: get universe-wide global variable Get universe-wide global variable. This is a common global that all CELL's can query with this instruction. RETURNS: ENERGY: set universe-wide global variable Set universe-wide global variable. This is a common global that all CELL's can write to with this instruction. RETURNS: ENERGY: get strain-wide global variable Get strain-wide global variable. Each strain has its own S0 global variable. This instruction allows the strain specific variable to
be queried.
This variable can be used by a swarm of organisms, from the same strain, to coordinate their activies.
RETURNS: ENERGY: set strain-wide global variable Set strain-wide global variable. Each strain has its own S0 global variable. This instruction allows the strain specific variable to
be written to.
This variable can be used by a swarm of organisms, from the same strain, to coordinate their activies.
RETURNS: ENERGY: total population Return the current population. RETURNS: The total population of the universe. This is the total number or organisms. ENERGY: strain population Return the current population of my strain. RETURNS: The population (number of organisms) for my strain. ENERGY: read keyboard Get the keyboard character being pressed. Or 0 if nothing is being pressed. RETURNS: Returns the ascii character code. Returns 0 if no key is pressed. ENERGY: KEY-PRESS MODE BITS: get mouse position Query the mouse position. The mouse position is a coordinate in the universe coordinate space. This position was set externally
by the user right clicking on a grid location when in 'MOUSE-POS' mode. See the KEY-PRESS instruction modes to configure
interrupts for the mouse position. RETURNS: Returns a coordinate in the universe which was set by the user clicking in the grid when in MOUSE-POS mode.
Returns (-1,-1) if no position set. ENERGY: compute vector distance Computes the maximum of ABS(x) and ABS(y). RETURNS: ENERGY:OMOVE
Usage: ( x y -- n )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 NOT USED NOT USED ROTATE
Usage: ( n -- r )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 use my location as the origin of rotation use the organism center as the origin of rotation 1 2 rotate in 45 degree units rotate in 90 degree units EAT
Usage: ( x y -- n )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 I cannot eat myself I can eat myself 1 2 I can eat cells from other strains I cannot eat cells from other strains 2 4 I can eat my own strain I cannot eat my own strain 3 8 I can be eaten I cannot be eaten 4 16 energy eaten is: cell energy + remainder (this bit is respected only if bits 5, 6, 7,8 are off) energy eaten is: cell energy 5 32 off use make-spore energy for maximum energy eaten 6 64 off use grow energy for maximum energy eaten 7 128 off use cell energy/2 + remainder 8 256 off use cell energy/3 + remainder 9 512 terminate cell eaten don't terminate cell eaten, unless energy per cell == 0 10 1024 do not interrupt me, when somebody EATS me interrupt me and bit 0 of the trap number shall be 1 11 2048 do not interrupt me, when somebody EATS me interrupt me and bit 1 of the trap number shall be 1 12 4096 do not interrupt me, when somebody EATS me interrupt me and bit 2 of the trap number shall be 1 SEND-ENERGY
Usage: (e x y -- rc)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i can give energy to other strains i cannot give energy to other strains 1 2 i can take energy from other strains i cannot take energy from other strains 2 4 i can give energy to spores i cannot give energy to spores 3 8 i can take energy from spores i cannot take energy from spores 4 16 do not interrupt me, when somebody gives energy to me interrupt me and bit 0 of the trap number shall be 1 5 32 do not interrupt me, when somebody gives energy to me interrupt me and bit 1 of the trap number shall be 1 6 64 do not interrupt me, when somebody gives energy to me interrupt me and bit 2 of the trap number shall be 1 7 128 do not interrupt me, when somebody takes energy from me interrupt me and bit 0 of the trap number shall be 1 8 256 do not interrupt me, when somebody takes energy from me interrupt me and bit 1 of the trap number shall be 1 9 512 do not interrupt me, when somebody take energy from me interrupt me and bit 2 of the trap number shall be 1 10 1024 can give energy only to adjacent neighbors can give energy across distances along normalized vector MAKE-SPORE
Usage: ( x y e -- r )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 NOT USED NOT USED 1 2 i can fertilize a spore i created i cannot fertilize a spore i created 2 4 i can fertilize a spore created by others i cannot fertilize a spore created by others 3 8 i can create a spore i cannot create a spore 4 16 i can fertilize a spore i cannot fertilize a spore 5 32 no energy is required to fertilize a spore minimum MAKE-SPORE Energy is required to fertilize a spore GROW
Usage: ( x y -- r )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 NOT USED NOT USED MAKE-ORGANIC
Usage: (x y e -- s)
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 NOT USED NOT USED GROW.CB
Usage: (x y cb -- r)
CSHIFT
Usage: (x y -- r)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 NOT USED NOT USED SPAWN
Usage: (x y e strain cb -- r)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 do not inherit cell register values inherit register values 1 2 num. protected code blocks follows new strain num. protected code blocks follows old strain 2 4 cannot spawn to a different strain can spawn to a different strain 3 8 mutate the program using my strain's mutations settings do not mutate the program 4 16 do not inherit data stack inherit N data stack items from top and bit-0 of N is 1 5 32 do not inherit data stack inherit N data stack items from top and bit-1 of N is 1 6 64 do not inherit data stack inherit N data stack items from top and bit-2 of N is 1 MAKE-BARRIER
Usage: (x y -- r)
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 allowed to create a barrier not allowed to create a barrier 1 2 allowed to clear a barrier not allowed to clear a barrier EXUDE
Usage: (value x y -- )BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i can exude on any surrounding square i cannot exude on any surrounding square, if it's occupied by anything 1 2 i can exude on any surrounding square i cannot exude on any surrounding square, if it's occupied by barrier 2 4 i can exude on any surrounding square, if it's occupied by cells from another organism i cannot exude on any surrounding square, if it's occupied by cell from another organism 3 8 i can exude on any surrounding square, if it's occupied by cell from another strain i cannot exude on any surrounding square, if it's occupied by cell from another strain 4 16 i can exude on a spore i cannot exude on a spore 5 32 i can exude on an organic block i cannot exude an an organic block SMELL
Usage: (x y -- value)LOOK
Usage: ( x y -- what dist )
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 when looking along a line any cells that belong to my organism are ignored when looking along a line any cells that belong to my organism stop the looking and report nothing 1 2 NOT USED NOT USED 2 4 when reporting a 'what' bit mask, don't include strain bits when reporting a 'what' bit mask, include strain bits 3 8 i am not invisible i am invisible NEAREST
Usage: ( mask -- x y )
FARTHEST
Usage: ( mask -- x y )
SIZE
Usage: (x y -- size dist)BIGGEST
Usage: (mask -- x y)
SMALLEST
Usage: (mask -- x y)
TEMPERATURE
Usage: (x y -- energy dist)
HOTTEST
Usage: (mask -- x y)
COLDEST
Usage: (mask -- x y)
MOOD
Usage: ( x y -- m )
0 0 MOOD ; returns -23
-1 0 MOOD ; returns 123
-1 1 MOOD ; returns 33
99 99 MOOD ; returns 0
MOOD!
Usage: ( m -- )
0 0 MOOD 1 + MOOD!
BROADCAST
Usage: ( m -- )BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 do not interrupt me, when somebody broadcasts a message to me interrupt me and bit 0 of the trap number shall be 1 1 2 do not interrupt me, when somebody broadcasts a message to me interrupt me and bit 1 of the trap number shall be 1 2 4 do not interrupt me, when somebody broadcasts a message to me interrupt me and bit 2 of the trap number shall be 1 SEND
Usage: ( m x y -- )
100 1 0 SEND ; set cell 2's message register to 100
-46812 0 1 SEND ; set cell 3's message register to -46812
0 0 0 SEND ; set out message register to 0.
12 -99 99 SEND ; does nothing because (-99, 99) is bogus
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 do not interrupt me, when somebody sends a message to me interrupt me and bit 0 of the trap number shall be 1 1 2 do not interrupt me, when somebody sends a message to me interrupt me and bit 1 of the trap number shall be 1 2 4 do not interrupt me, when somebody sends a message to me interrupt me and bit 2 of the trap number shall be 1 RECV
Usage: ( -- m )SHOUT
Usage: (m -- r)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i can shout thru myself i cannot shout thru myself 1 2 i can shout at cells that belong to my strain i cannot shout at cells that belong to my strain 2 4 i can shout at cells that belong to OTHER strains i cannot shout at cells that belong to OTHER strains 3 8 other strains can shout at me other strains cannot shout at me 4 16 do not interrupt me, when somebody shouts a message to me interrupt me and bit 0 of the trap number shall be 1 5 32 do not interrupt me, when somebody shouts a message to me interrupt me and bit 1 of the trap number shall be 1 6 64 do not interrupt me, when somebody shouts a message to me interrupt me and bit 2 of the trap number shall be 1 SAY
Usage: (m x y -- dist)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i can speak thru myself i cannot speak thru myself 1 2 i can speak at cell that belong to my organism i cannot speak at cell that belong to my organism 2 4 i can speak at cells that belong to my strain i cannot speak at cells that belong to my strain 3 8 i can speak at cell that belong to OTHER strains i cannot speak at cell that belong to OTHER strains 4 16 other strains can speak at me other strains cannot speak at me 5 32 do not interrupt me, when somebody speaks a message to me interrupt me and bit 0 of the trap number shall be 1 6 64 do not interrupt me, when somebody speaks a message to me interrupt me and bit 1 of the trap number shall be 1 7 128 do not interrupt me, when somebody speaks a message to me interrupt me and bit 2 of the trap number shall be 1 LISTEN
Usage: (x y -- mood dist)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 when listening along a line any cells that belong to my organism are ignored when listening along a line any cells that belong to my organism stop the listing process and report nothing READ
Usage: (x y cb cbme -- rc)
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i cannot read from myself i can read from myself 1 2 i can read from cells/spores that belong to my strain i cannot read from cells/spores that belong to my strain 2 4 i cannot read from cells/spores that belong to OTHER strains i can read from cells/spores that belong to OTHER strains 3 8 i can read from spores i cannot read from spores 4 16 i can read from cells i cannot read from cells 5 32 other strains can read from me other strains cannot read from me 6 64 mutate the code block being read with my strain's mutation settings. do not mutate the code block being read WRITE
Usage: (x y cb cbme -- rc)
BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 i cannot write to myself i can write to myself 1 2 i can write to cells/spores that belong to my strain i cannot write to cells/spores that belong to my strain 2 4 i cannot write to cells/spores that belong to OTHER strains i can write to cells/spores that belong to OTHER strains 3 8 i can write to spores i cannot write to spores 4 16 i cannot write to cells i can write to cells 5 32 other strains can write to me other strains cannot write to at me 6 64 mutate the code block being written with my strain's mutation settings. do not mutate the code block being written 7 128 do not interrupt me, when somebody writes to me interrupt me and bit 0 of the trap number shall be 1 8 256 do not interrupt me, when somebody writes to me interrupt me and bit 1 of the trap number shall be 1 9 512 do not interrupt me, when somebody writes to me interrupt me and bit 2 of the trap number shall be 1 ENERGY
Usage: ( -- e )AGE
Usage: ( -- a )NUM-CELLS
Usage: ( -- n )HAS-NEIGHBOR
Usage: ( x y -- r )
NEIGHBORS
Usage: ( -- mask)
GPS
Usage: ( -- x y)G0
Usage: ( -- value)G0!
Usage: (value -- )S0
Usage: ( -- value)S0!
Usage: (value -- )POPULATION
Usage: ( -- pop)POPULATION.S
Usage: ( -- pop)KEY-PRESS
Usage: ( -- key)BIT MASK Meaning when bit is 0 Meaning when bit is 1 0 1 do not interrupt me, when KEY-PRESS changes interrupt me on KEY-PRESS change, and bit 0 of the trap number shall be 1 1 2 do not interrupt me, when KEY-PRESS changes interrupt me on KEY-PRESS change, and bit 1 of the trap number shall be 1 2 4 do not interrupt me, when KEY-PRESS changes interrupt me on KEY-PRESS change, and bit 2 of the trap number shall be 1 3 8 do not interrupt me, when MOUSE-POS changes interrupt me on MOUSE-POS change, and bit 0 of the trap number shall be 1 4 16 do not interrupt me, when MOUSE-POS changes interrupt me on MOUSE-POS change, and bit 1 of the trap number shall be 1 5 32 do not interrupt me, when MOUSE-POS changes interrupt me on MOUSE-POS change, and bit 2 of the trap number shall be 1 MOUSE-POS
Usage: ( -- x y)DIST
Usage: (x y -- dist)